
Ulf Schmitz
- ulf.schmitz@jcu.edu.au
 https://orcid.org/0000-0001-5806-4662
https://orcid.org/0000-0001-5806-4662- Associate Professor, Bioinformatics
Projects
10
Publications
11
Awards
8
Contact Details
- 0747816265
- ulf.schmitz@jcu.edu.au
- https://jcu.me/ulf.schmitz
-
The Science Place
1 James Cook Drive
Townsville QLD 4811
AUSTRALIA
Biography
Ulf is a computational biologist with training in bioinformatics and systems biology and more than 15 years of experience in analysing post-transcriptional gene regulation. His research interests focus on computational RNA biology and Systems Medicine.
Ulf completed his PhD at the University of Rostock (Germany). In 2015, he joined the Centenary Institute in Sydney as a post-doctoral Research Officer in the Gene and Stem Cell Therapy Program. He later established the Computational Biomedicine Lab at the Centenary. He also holds an appointment as Conjoint Senior Lecturer at the University of Sydney (Faculty of Medicine & Health).
Ulf and his team develop integrative workflows combining various computational disciplines with experimentation to address questions around non-coding RNAs, post-transcriptional gene regulation, and cancer biology. He has published 65 research articles, 13 book chapters, and 4 books on this topic.
Available HDR Student Projects
Blood-Based Surveillance for HPV-Associated Oropharyngeal Cancer
Mathematical Modelling of Alternative Splicing and Transcriptomic Complexity
Mathematical Modelling of Cell-Fate Determination
Computational Biomedicine Lab
In the Computational Biomedicine Lab at James Cook University computational biologists work hand-in-hand with molecular biologists integrating in silico, in vitro and in vivo approaches in advanced interdisciplinary research. We develop integrative systems medicine workflows, tools, and databases to identify and study interactions between RNAs and other molecules, functional mechanisms of gene regulation, cell signalling pathways, and gene regulatory networks.
Our goal is to achieve a deeper understanding of processes involved in disease emergence and progression, with a focus on cancer biology, and possible avenues for therapeutic interventions and optimised treatment schedules. Most projects involve the analysis of experimental high-throughput data (incl. (sc)RNA-seq, NanoString, DNA Methyl-Seq, ChIP-Seq, CLIP-seq, and more). Moreover, we use various mathematical formalisms to model the effects of environmental stimuli or pathogenic events on molecular interaction networks and signalling pathways. Using machine learning and computer simulation approaches, we predict disease progression and outcome as well as the effect of therapeutic agents on cellular survival and resistance mechanisms.
Current Research Projects
Transcriptomic Complexity In Single Cells
Most single-cell analysis focuses on gene expression, but alternative splicing creates multiple transcript isoforms from each gene - adding layers of regulatory complexity that current methods miss. We develop computational frameworks for measuring transcriptomic complexity in single-cell sequencing data. See explainer video for our recently published software ScIsoX here?
Decoding Genetic Complexity
MicroRNAs as Cancer Biomarkers
MicroRNAs are small molecules that play an important role in regulating gene expression. We have been exploring the potential of microRNAs as cancer biomarkers, as regulators of cancer genes and their role in immune cells. For more information about our work see recent media coverage:
Intron retention regulation in haematopoietic cells
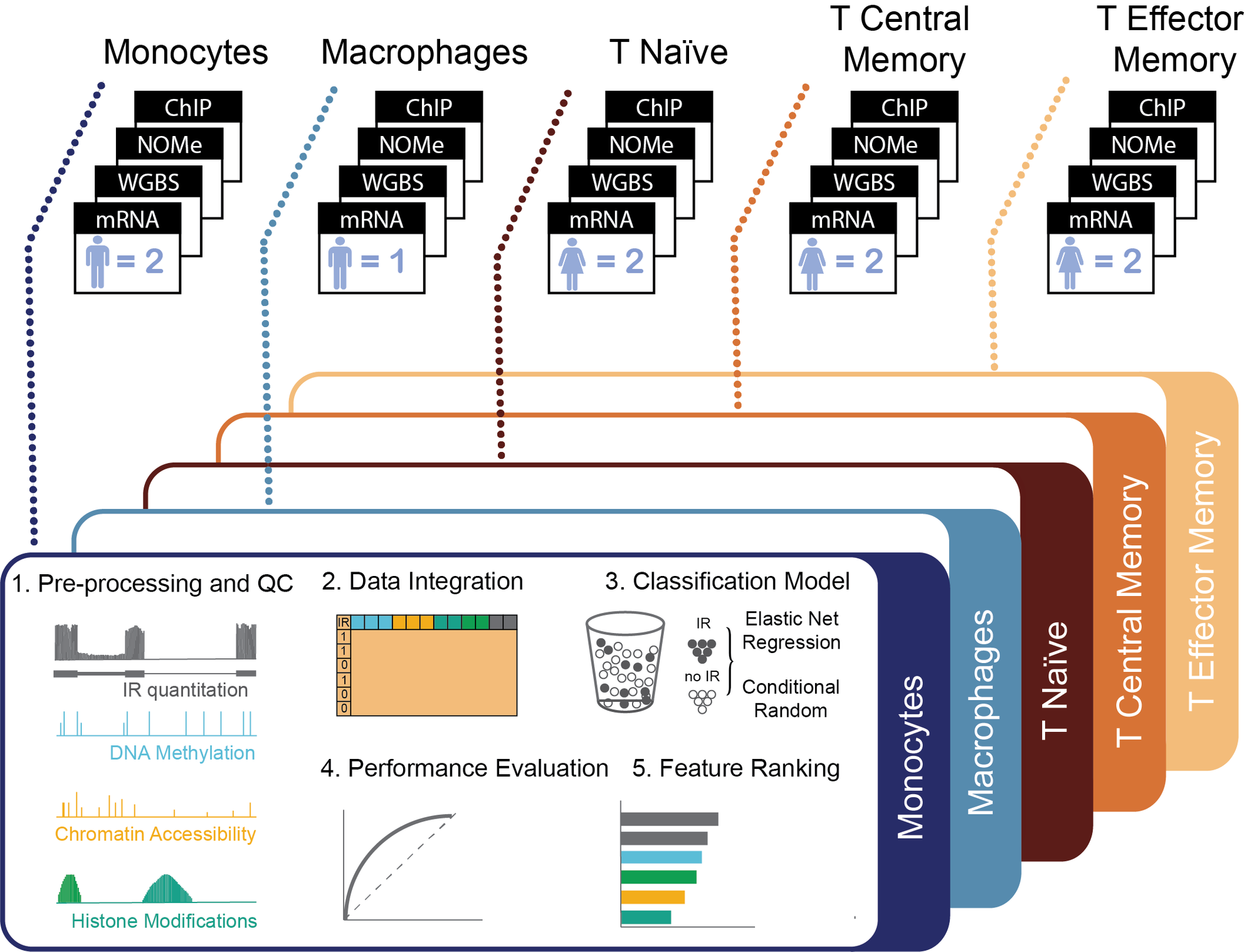
There is growing evidence that alternative splicing, including intron retention (IR), is regulated on at least two levels: locally, through a network of interacting trans-acting splicing regulators with cis-acting regulatory elements, and globally, through the chromatin structure. In this project, we explore a global regulation of IR in haematopoietic cells – we use statistical analysis to better understand IR regulation using information derived from the epigenetic factors that govern chromatin organisation, namely nucleosome assembly, DNA methylation, and histone modifications. The main challenge here is to integrate multiple layers of ‘-omics’ into a single computational model which produces biologically interpretable results. Find out more: Petrova et al, Nucleic Acids Research 2022
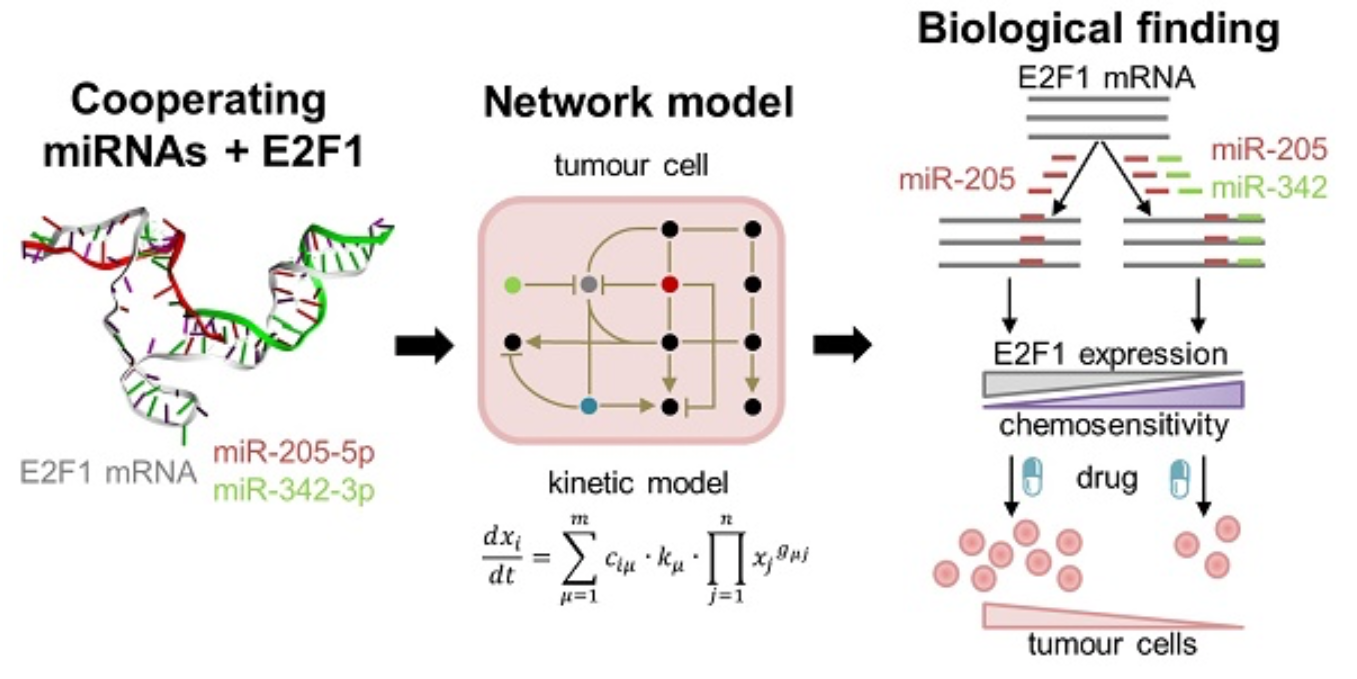
Cooperating microRNAs for cancer therapy
In this project, we use a systems medicine approach to find new avenues for overcoming chemotherapy resistance in aggressive tumour cells. Our results suggest that pairs of cooperating microRNAs could be used as potential RNA therapeutics to reduce E2F1-related chemoresistance. Find out more: Lai X, Gupta SK, Schmitz U et al, Theranostics 2018
Alternative splicing and the epigenome in Chronic Myeloid Leukaemia
In this project, we are investigating gene regulatory processes in leukaemia. The third biggest cause of cancer death in all Australians is blood cancers (leukaemia), which are diagnosed 35 times each day. Using a multi-omics approach, we examine alternative splicing and epigenetic changes in blood samples from chronic myeloid leukaemia (CML) patients before and after treatment with tyrosine kinase inhibitors. Find out more: Schmitz et al, Cancers 2020

Cross-talk between post-transcriptional gene regulation mechanisms
Combining computational predictions and in vitro assays, we elucidate the cross-talk between post-transcriptional gene regulation mechanisms. More specifically, we focus on interactions between microRNA molecules and intron-retaining mRNA transcripts and try to identify networks of interconnected gene regulation. Using a systems biology approach we will model competitive post-transcriptional gene regulation through iterative cycles of time-course experiments and model simulations.

Portable long-read sequencing to diagnose Chronic Kidney Disease
With the help of new seed funding from the Tropical Australian Academic Health Centre, we trial a portable, rapid, and targeted genomic test to find genetic variants that predispose people towards Chronic Kidney Disease. For more information see recent media coverage:






